Arabidopsis thaliana, L.
|
publication ID |
https://doi.org/10.1016/j.phytochem.2020.112511 |
|
DOI |
https://doi.org/10.5281/zenodo.8301829 |
|
persistent identifier |
https://treatment.plazi.org/id/442187AF-FFCC-FFD2-2869-F8D5FA520388 |
|
treatment provided by |
Felipe |
|
scientific name |
Arabidopsis thaliana |
| status |
|
2.1. The majority of A. thaliana View in CoL View at ENA defensins belongs to class I defensins
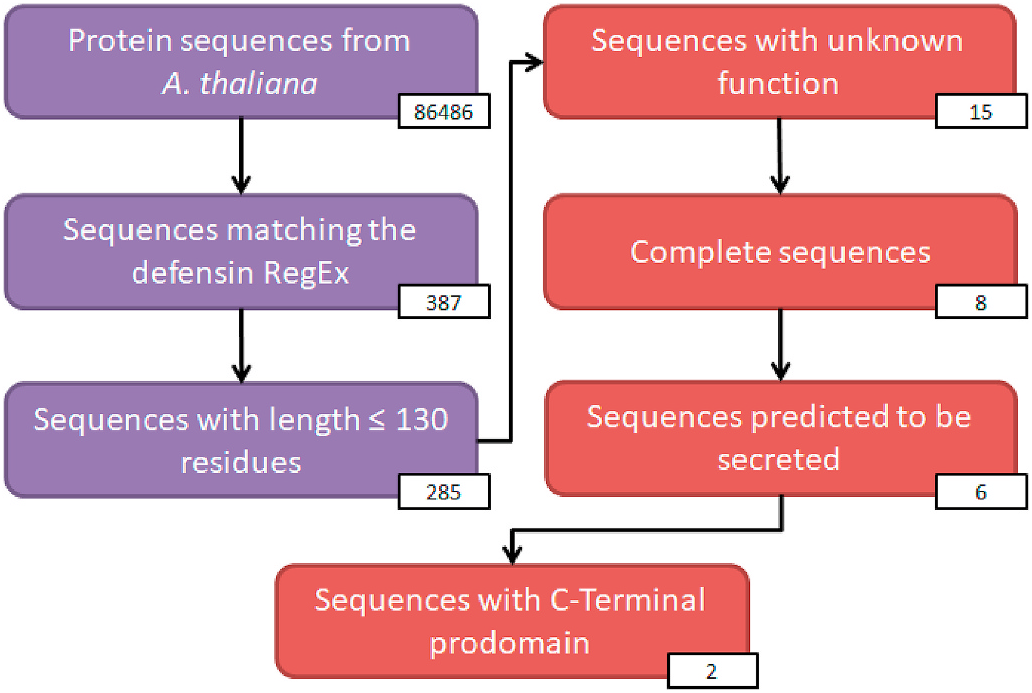
In order to identify class II defensin sequences, we designed a semiautomatic pipeline ( Fig. 1 View Fig ). For that, initially all proteins from the A. thaliana Uniprot database were downloaded. The dataset consisted of 86,486 sequences ( March 2017). From this dataset, 387 sequences were retrieved by using regular expression (RegEx) search (step 2, Fig. 1 View Fig ). From these, 285 had up to 130 amino acid residues (step 3, Fig. 1 View Fig ). This criterion allows eventual larger C-terminal prodomains to be identified. Then, we used a PERL script to select the sequences with the following flags: hypothetical, unknown, unnamed and/or uncharacterized (step 4, Fig. 1 View Fig ), resulting in 15 sequences. From 15 sequences, seven were incomplete and therefore were discarded, (step 5, Fig. 1 View Fig ). From the remaining sequences, two sequences without signal peptide or with transmembrane domains were discarded (step 6, Fig. 1 View Fig ). Finally, from six remaining sequences, two sequences with a potential C-Terminal prodomain were selected, with accession codes A7 REG2 About REG and A7 REG4 About REG .
No known copyright restrictions apply. See Agosti, D., Egloff, W., 2009. Taxonomic information exchange and copyright: the Plazi approach. BMC Research Notes 2009, 2:53 for further explanation.
|
Kingdom |
|
|
Phylum |
|
|
Class |
|
|
Order |
|
|
Family |
|
|
Genus |